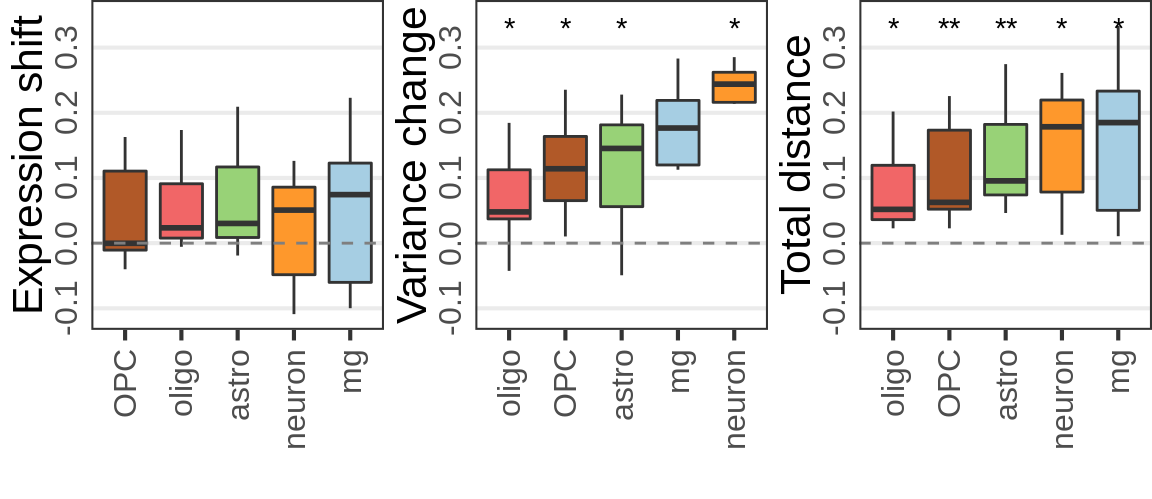
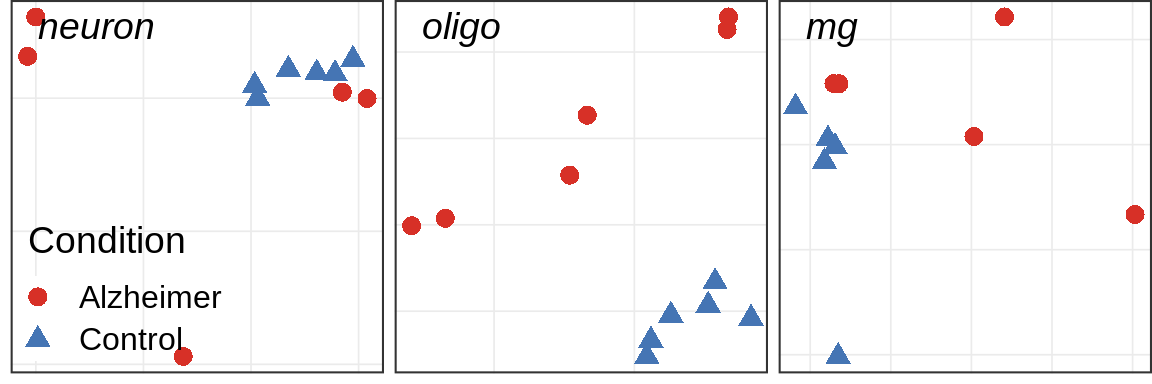
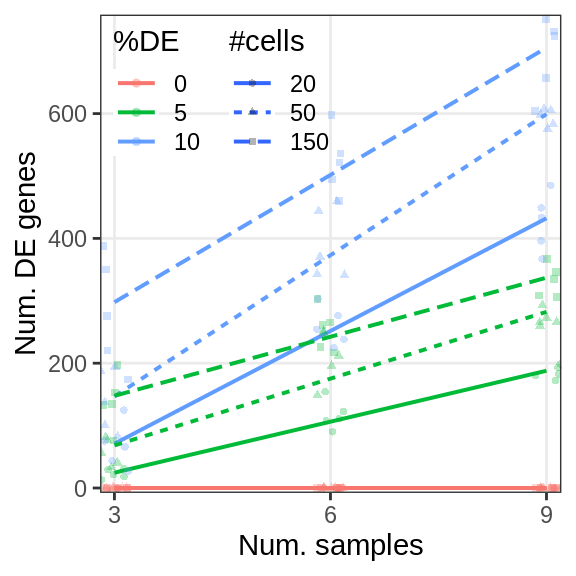
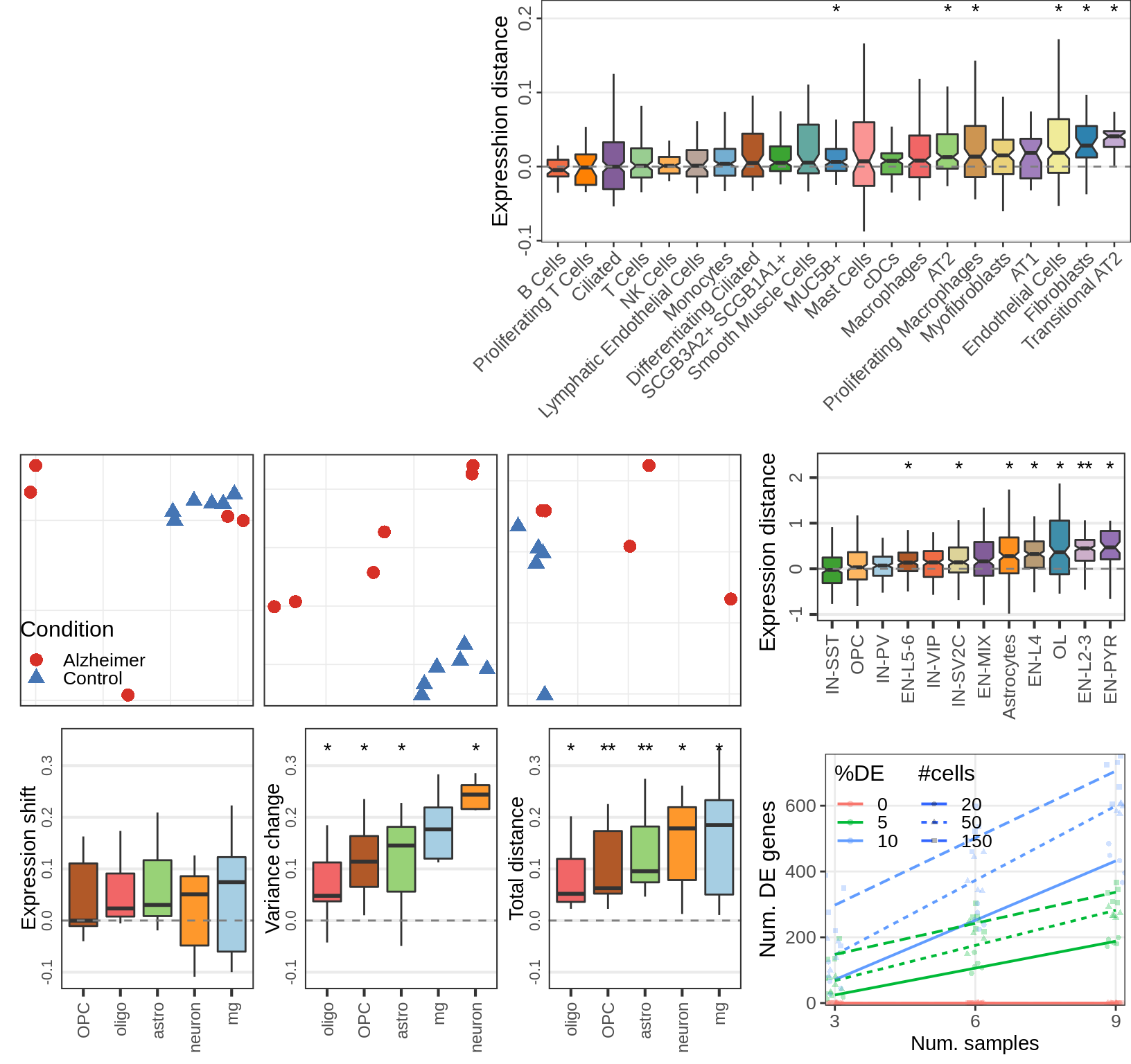
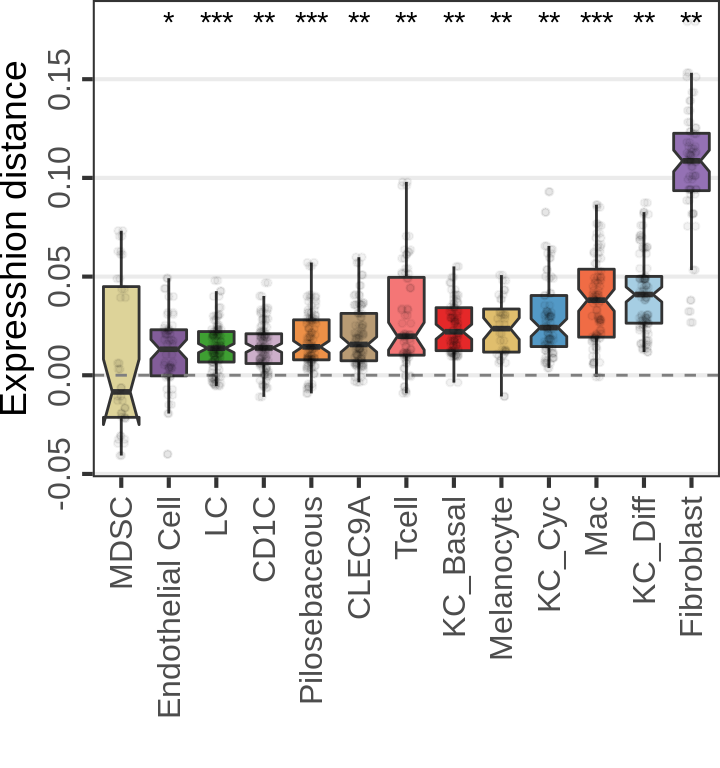
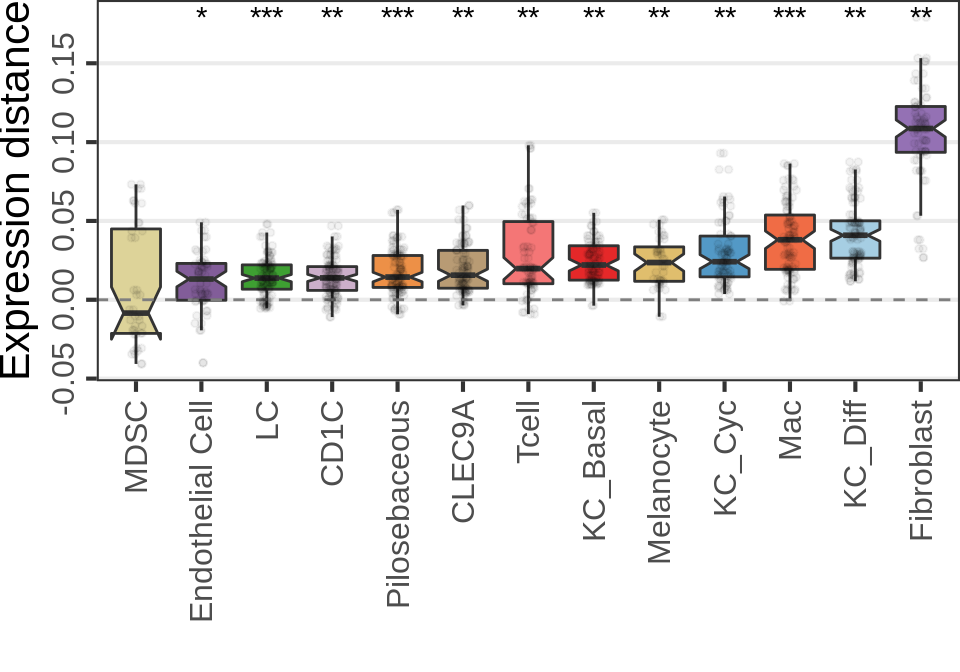
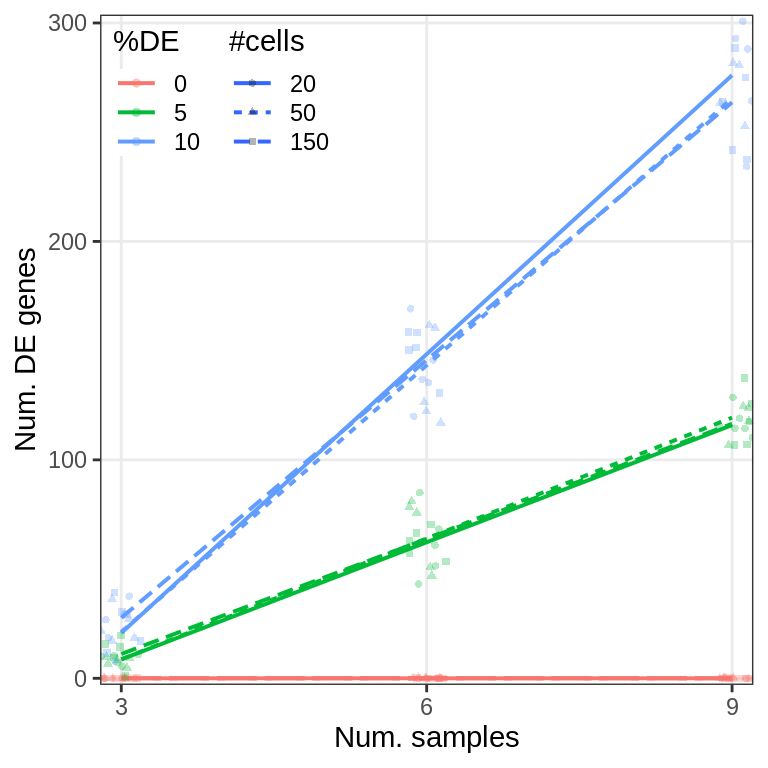
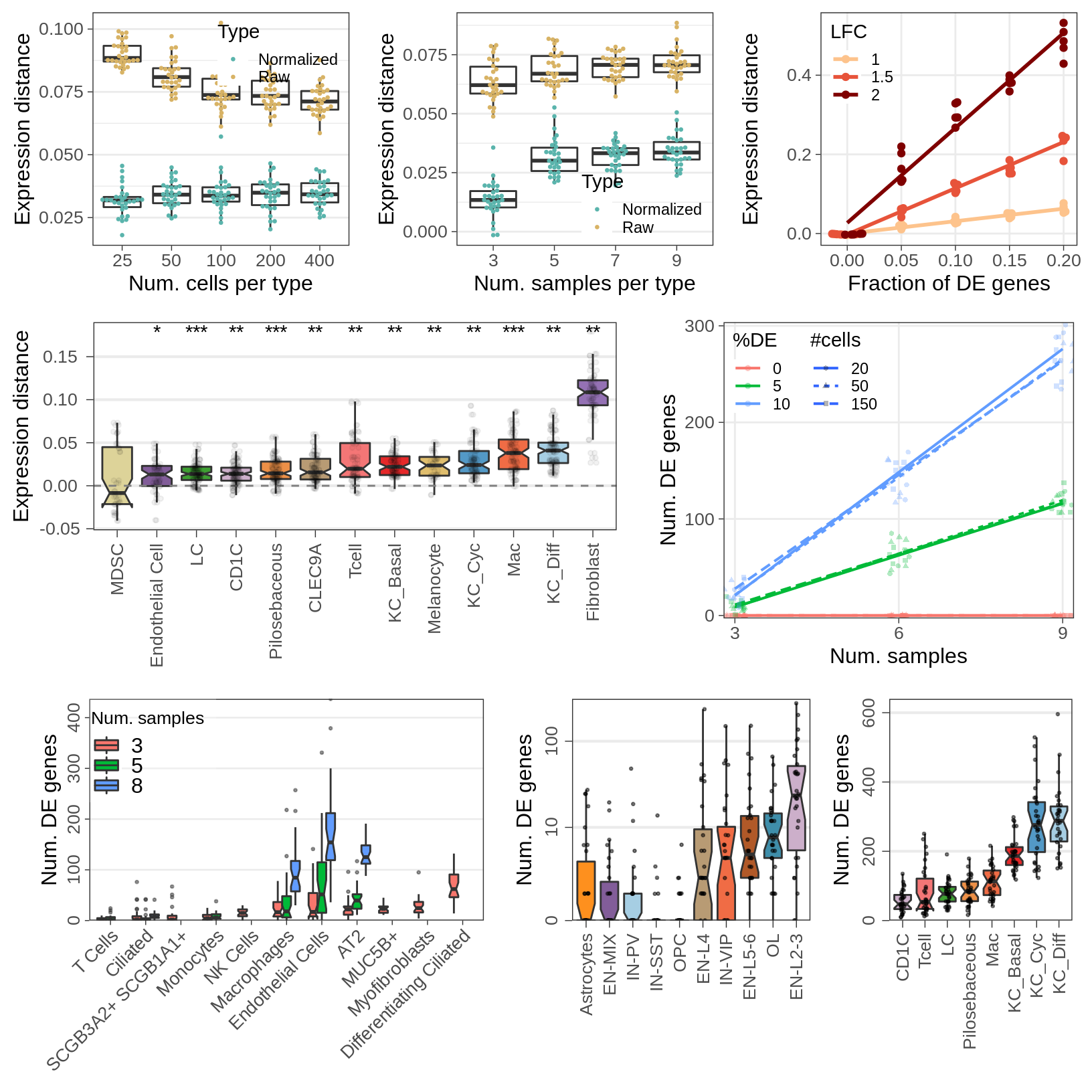
Covariate simulations
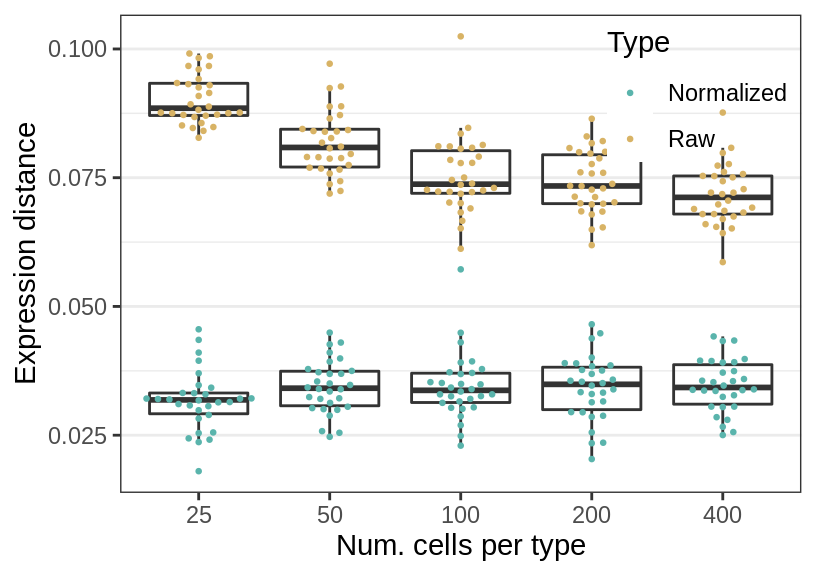
Num. cells
sim_nc <- generateSims(sce, n.cells=c(25, 50, 100, 200, 400), n.samples=8,
de.frac=0.1, lfc=1, n.cores=N_CORES, n.repeats=30)
75 75 75 15 15 15
cao_sim_nc <- cacoaFromSim(sim_nc, n.cores=N_CORES)
cao_sim_nc$estimateExpressionShiftMagnitudes(
verbose=TRUE, n.permutations=N_PERMUTS, min.samp.per.type=1, n.cores=N_CORES
)
sens_df_adj <- cao_sim_nc$test.results$expression.shifts %$%
{list(dist.df=prepareExpressionDistDf(dists.per.type, params=sim_nc$params))} %>%
prepareSensitivityDf(params=sim_nc$params, covar.name="nc") %>% mutate(Type="Normalized")
p_dists <- cao_sim_nc$test.results$expression.shifts$p.dist.info %>%
lapply(cacoa:::subsetDistanceMatrix, sample.groups=cao_sim_nc$sample.groups,
cross.factor=TRUE, build.df=FALSE)
sens_df_raw <- list(dist.df=prepareExpressionDistDf(p_dists, params=sim_nc$params)) %>%
prepareSensitivityDf(params=sim_nc$params, covar.name="nc") %>% mutate(Type="Raw")
gg_nc <- ggplot(rbind(sens_df_raw, sens_df_adj), aes(x=nc, y=value)) +
geom_boxplot(aes(fill=Type), alpha=0.0, position="identity") +
ggbeeswarm::geom_beeswarm(aes(color=Type), cex=1.75, size=0.5) +
guides(fill="none") +
labs(x="Num. cells per type", y="Expression distance") +
cao_sim_nc$plot.theme + theme(panel.grid.major.x=element_blank()) +
theme_legend_position(c(1, 1)) +
theme(legend.background=element_blank()) +
scale_color_manual(values=c("#5ab4ac", "#d8b365"))
gg_nc
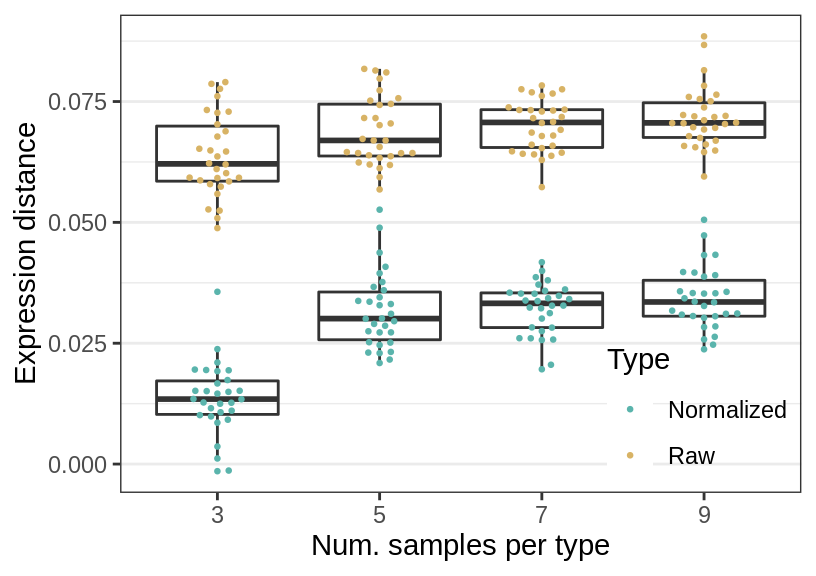
Num. samples
sim_ns <- generateSims(sce, n.cells=300, n.samples=c(3, 5, 7, 9),
de.frac=0.1, lfc=1, n.cores=N_CORES, n.repeats=30)
75 75 18 18 18 18
cao_sim_ns <- cacoaFromSim(sim_ns, n.cores=N_CORES)
cao_sim_ns$estimateExpressionShiftMagnitudes(
verbose=TRUE, n.permutations=N_PERMUTS, min.samp.per.type=1, n.cores=N_CORES
)
sens_df_adj <- cao_sim_ns$test.results$expression.shifts %$%
{list(dist.df=prepareExpressionDistDf(dists.per.type, params=sim_ns$params))} %>%
prepareSensitivityDf(params=sim_ns$params, covar.name="nc") %>% mutate(Type="Normalized")
p_dists <- cao_sim_ns$test.results$expression.shifts$p.dist.info %>%
lapply(cacoa:::subsetDistanceMatrix, sample.groups=cao_sim_ns$sample.groups, cross.factor=TRUE, build.df=FALSE)
sens_df_raw <- list(dist.df=prepareExpressionDistDf(p_dists, params=sim_ns$params)) %>%
prepareSensitivityDf(params=sim_ns$params, covar.name="nc") %>% mutate(Type="Raw")
gg_ns <- ggplot(rbind(sens_df_raw, sens_df_adj), aes(x=ns, y=value)) +
geom_boxplot(aes(fill=Type), alpha=0.0, position="identity") +
ggbeeswarm::geom_beeswarm(aes(color=Type), cex=1.75, size=0.5) +
guides(fill="none") +
labs(x="Num. samples per type", y="Expression distance") +
cao_sim_ns$plot.theme + theme(panel.grid.major.x=element_blank()) +
theme_legend_position(c(1, 0)) +
theme(legend.background=element_blank()) +
scale_color_manual(values=c("#5ab4ac", "#d8b365"))
gg_ns
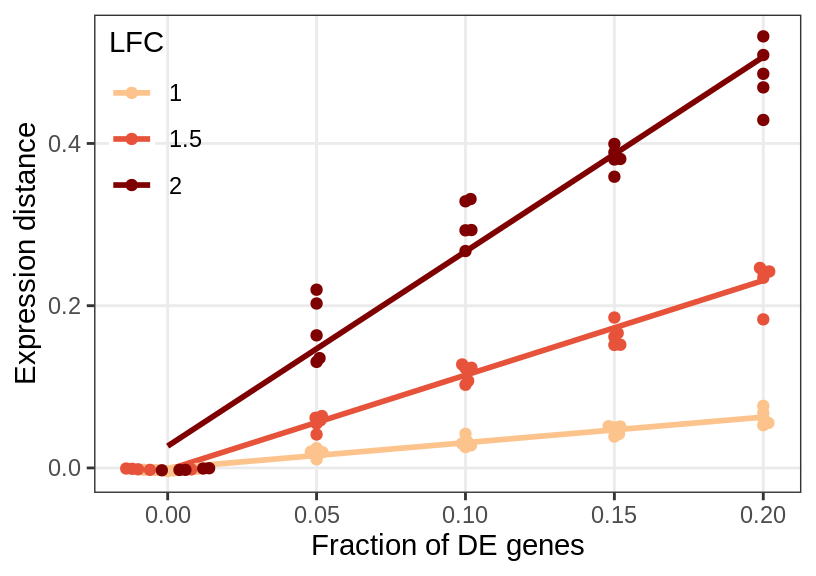
Sensitivity
de_fracs <- seq(0.0, 0.2, by=0.05)
sim_sens <- generateSims(sce, n.cells=300, n.samples=8, de.frac=de_fracs,
lfc=c(1, 1.5, 2), n.cores=N_CORES, n.repeats=5)
75 25 25 25 5 5
cao_sim_sens <- cacoaFromSim(sim_sens, n.cores=N_CORES)
cao_sim_sens$estimateExpressionShiftMagnitudes(
verbose=TRUE, n.permutations=N_PERMUTS, min.samp.per.type=1, n.cores=N_CORES
)
sens_df_adj <- cao_sim_sens$test.results$expression.shifts %$%
{list(dist.df=prepareExpressionDistDf(dists.per.type, params=sim_sens$params), pvalues=padjust)} %>%
prepareSensitivityDf(params=sim_sens$params)
gg_sens <- ggplot(sens_df_adj, aes(x=de.frac, y=value, color=lfc)) +
geom_smooth(se=0, method=MASS::rlm, formula=y~x) +
ggbeeswarm::geom_beeswarm() +
guides(fill="none") +
labs(x="Fraction of DE genes", y="Expression distance", color="LFC") +
scale_x_continuous(breaks=de_fracs) +
theme_legend_position(c(0, 1)) +
theme(legend.background=element_blank(), panel.grid.minor=element_blank()) +
scale_color_manual(values=brewerPalette("OrRd", rev=FALSE)(4)[2:4])
gg_sens
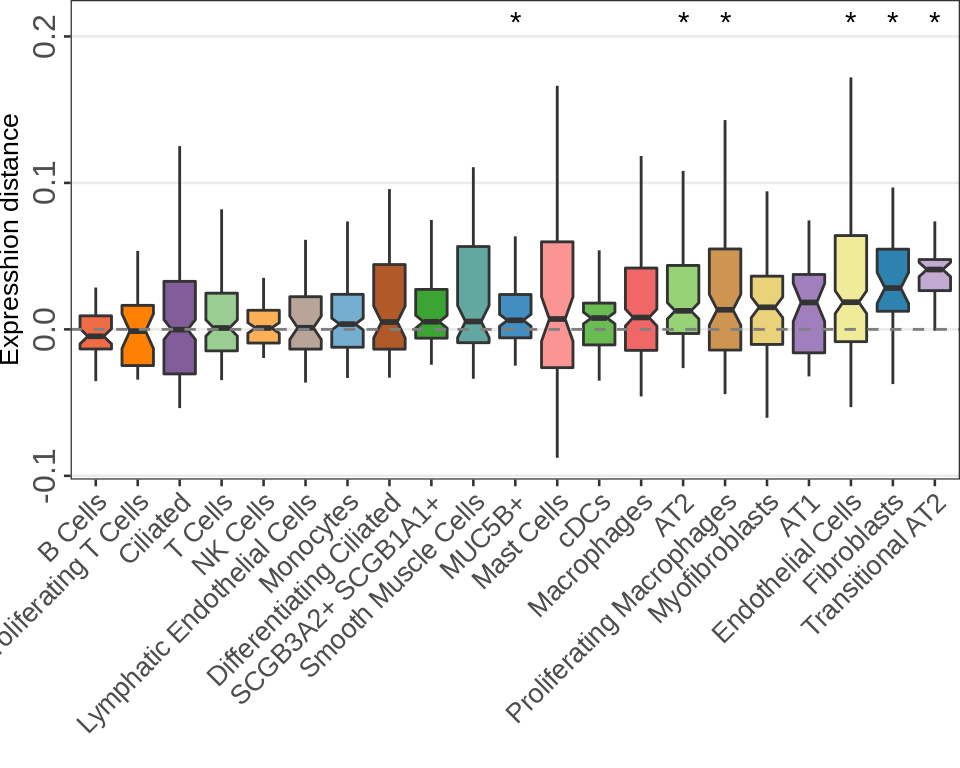
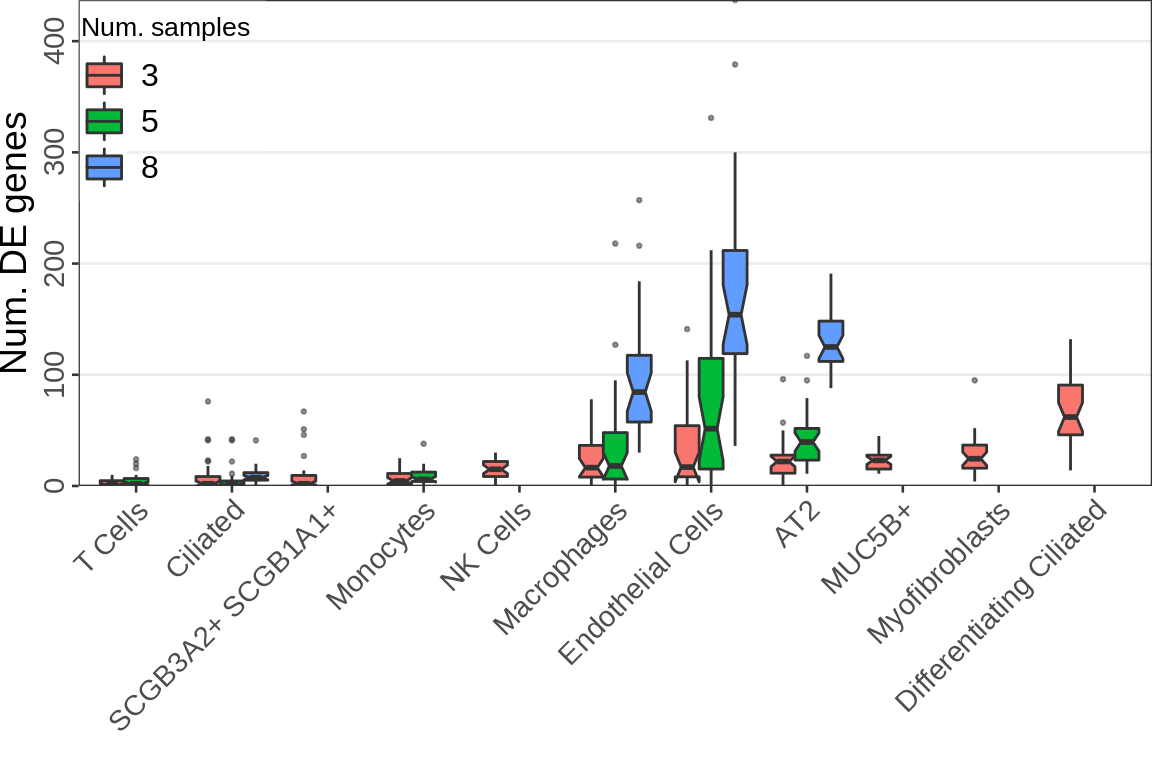
Real data num. DE
PF:
n_cells <- 30
for (ns in c(3, 5, 8)) {
cao_pf$estimateDEPerCellType(
independent.filtering=TRUE, test='DESeq2.Wald', name=paste0("de.fix.samples", ns),
resampling.method='fix.samples', n.cores=N_CORES, n.resamplings=N_RESAMPLES,
fix.n.samples=ns, n.cells.subsample=n_cells, verbose=TRUE
)
}
n_de_df <- c(3, 5, 8) %>% {setNames(cao_pf$test.results[paste0("de.fix.samples", .)], .)} %>%
lapply(function(tr) {
lapply(tr, `[[`, 'subsamples') %>% .[sapply(., length) > 0] %>%
lapply(function(r) tibble(value=sapply(r, function(df) sum(df$padj <= 0.05, na.rm=TRUE)))) %>%
bind_rows(.id="Type")
}) %>% bind_rows(.id="Num. samples")
sdf <- n_de_df %>% filter(`Num. samples` == 3) %>% group_by(Type) %>%
summarise(med=median(value)) %>% arrange(med)
n_de_df %<>% mutate(Type=factor(Type, levels=sdf$Type))
gg_nde_pf <- ggplot(n_de_df, aes(x=Type, y=value, fill=`Num. samples`)) +
geom_boxplot(position=position_dodge(preserve="single"), notch=TRUE, outlier.alpha=0.5, outlier.size=0.5) +
labs(x="", y="Num. DE genes") +
cao_pf$plot.theme + theme(
axis.text.x=element_text(angle=45, vjust=1, hjust=1, size=11),
axis.text.y=element_text(angle=90, hjust=0.5, size=11),
axis.title.y=element_text(size=14),
panel.grid.major.x=element_blank(), panel.grid.minor=element_blank()
) + theme_legend_position(c(0, 1)) + scale_y_continuous(expand=c(0, 0))
gg_nde_pf
# cacoa:::plotMeanMedValuesPerCellType
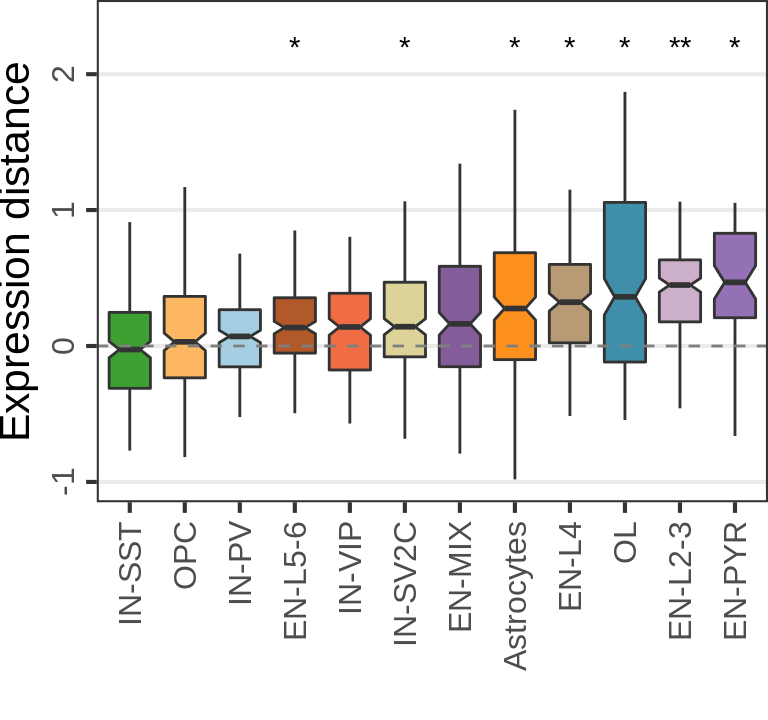
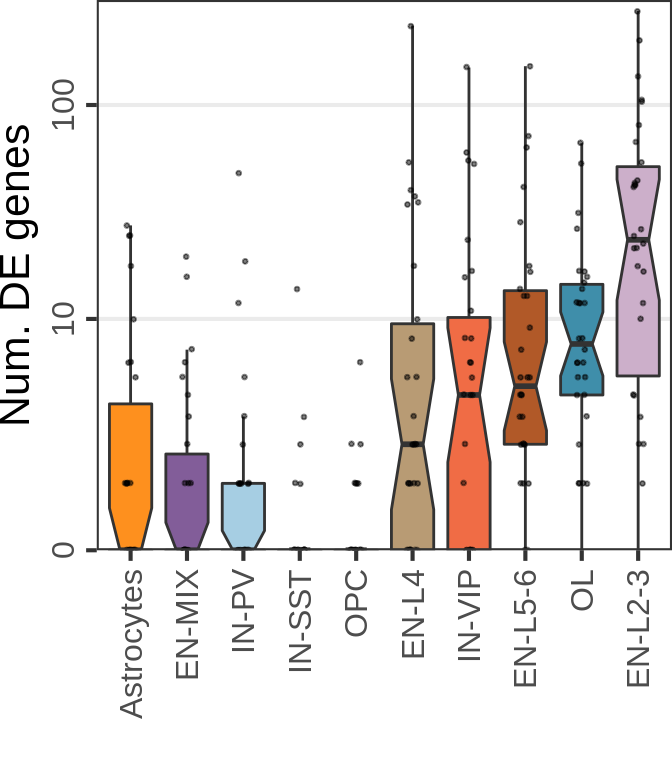
MS:
n_cells <- 30
n_samples <- 6
cao_ms$estimateDEPerCellType(
independent.filtering=TRUE, test='DESeq2.Wald', name="de.fix.samples",
resampling.method='fix.samples', n.cores=N_CORES, n.resamplings=N_RESAMPLES,
fix.n.samples=n_samples, n.cells.subsample=n_cells, verbose=TRUE
)
gg_nde_ms <- cao_ms$plotNumberOfDEGenes(
name="de.fix.samples", type="box", show.resampling.results=TRUE, jitter.alpha=0.5,
show.jitter=TRUE, jitter.size=0.5, y.offset=1
) + scale_y_log10(labels=c(0, 10, 100), breaks=c(1, 11, 101), expand=c(0, 0),
limits=c(1, 300), name="Num. DE genes")
gg_nde_ms
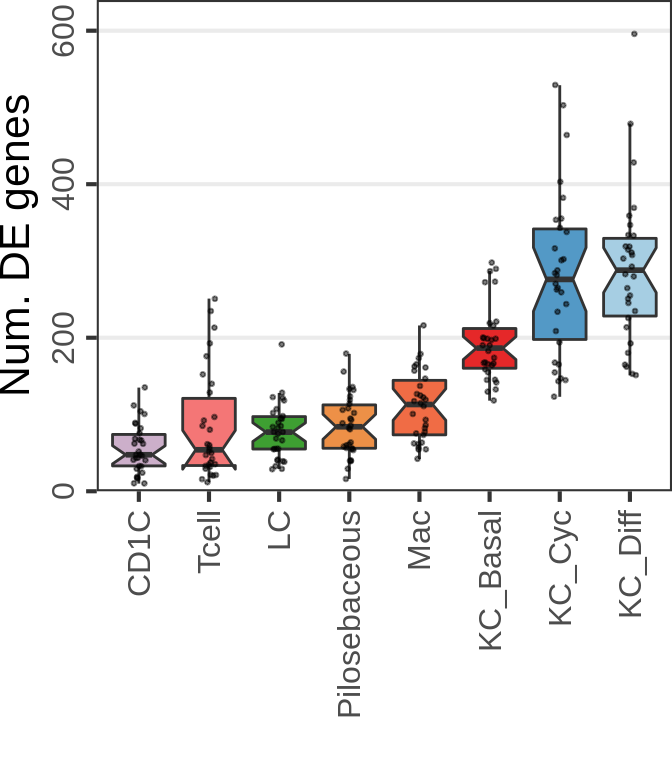
SCC:
n_cells <- 20
n_samples <- 4
cao_scc$estimateDEPerCellType(
independent.filtering=TRUE, test='DESeq2.Wald', name="de.fix.samples",
resampling.method='fix.samples', n.cores=N_CORES, n.resamplings=N_RESAMPLES,
fix.n.samples=n_samples, n.cells.subsample=n_cells, verbose=TRUE
)
gg_nde_scc <- cao_scc$plotNumberOfDEGenes(
name="de.fix.samples", type="box", show.resampling.results=TRUE, jitter.alpha=0.5,
show.jitter=TRUE, jitter.size=0.5
) + scale_y_continuous(limits=c(0, 640), expand=c(0, 0), name="Num. DE genes")
gg_nde_scc